issues
9 rows where user = 703554 sorted by updated_at descending
This data as json, CSV (advanced)
Suggested facets: comments, created_at (date), updated_at (date), closed_at (date)
| id | node_id | number | title | user | state | locked | assignee | milestone | comments | created_at | updated_at ▲ | closed_at | author_association | active_lock_reason | draft | pull_request | body | reactions | performed_via_github_app | state_reason | repo | type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 267628781 | MDU6SXNzdWUyNjc2Mjg3ODE= | 1650 | Low memory/out-of-core index? | alimanfoo 703554 | open | 0 | 17 | 2017-10-23T11:13:06Z | 2023-08-29T11:43:19Z | CONTRIBUTOR | Has anyone considered implementing an index for monotonic data that does not require loading all values into main memory? Motivation: We have data where first dimension can be length ~100,000,000, and coordinates for this dimension are stored as 32-bit integers. Currently if we used a pandas Index this would cast to 64-bit integers, and the index would require ~1GB RAM. This isn't enormous, but isn't negligible for people working on modest computers. Our use cases are simple, typically we only ever need to locate a slice of this dimension from a pair of coordinates, i.e., we only need to do binary search (bisect) on the coordinates. To achieve binary search in fact there is no need at all to load the coordinate values into memory, they could be left on disk (e.g., in HDF5 or Zarr dataset) and still achieve perfectly adequate performance for our needs. This is of course also relevant to pandas but thought I'd post here as I know there have been some discussions about how to handle indexes when working with larger datasets via dask. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/1650/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 1704950804 | I_kwDOAMm_X85ln3wU | 7833 | Slow performance of concat() | alimanfoo 703554 | closed | 0 | 3 | 2023-05-11T02:39:36Z | 2023-06-02T14:36:12Z | 2023-06-02T14:36:12Z | CONTRIBUTOR | What is your issue?In attempting to concatenate many datasets along a large dimension (total size ~100,000,000) I'm finding very slow performance, e.g., tens of seconds just to concatenate two datasets. With some profiling, I find all the time is being spend in this list comprehension: I don't know exactly what's going on here, but it doesn't look right - e.g., if the size of the dimension to be concatenated is large, this list comprehension can run millions of loops, which doesn't seem related to the intended behaviour. Sorry I don't have an MRE for this yet but please let me know if I can help further. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/7833/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 1558347743 | I_kwDOAMm_X85c4n_f | 7478 | Refresh list of backends (engines)? | alimanfoo 703554 | closed | 0 | 1 | 2023-01-26T15:41:24Z | 2023-03-31T15:14:58Z | 2023-03-31T15:14:58Z | CONTRIBUTOR | What is your issue?If a user is working in an environment where zarr is not initially installed (e.g., colab), then imports xarray, the list of available backends (engines) will not include zarr, as expected. If the user subsequently installs zarr, then tries to open a zarr dataset via
This can also be triggered in a more subtle way, because importing other packages like pandas will also trigger xarray to inspect and then fix this list of available engines. Obviously this can be resolved by instructing users to restart their kernel after any install commands, or by telling them to always run install commands before any imports. But I was wondering if there is a way to tell xarray to refresh its information about available engines? I have been able to hack it by running:
...but maybe there is a better way? Thanks :heart: |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/7478/reactions",
"total_count": 2,
"+1": 2,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 621078539 | MDU6SXNzdWU2MjEwNzg1Mzk= | 4079 | Unnamed dimensions | alimanfoo 703554 | open | 0 | 12 | 2020-05-19T15:33:48Z | 2023-02-18T16:51:14Z | CONTRIBUTOR | I'd like to build an xarray dataset with a couple of dozen variables. All of these variables share a common first dimension, which I want to name. Some of the arrays have other dimensions, but none of those dimensions are in common with any other arrays, and I don't want to ever refer to them by name. Currently IIUC xarray requires that all dimensions are named, which forces me to invent names for all these other dimensions. This ends up with a dataset with lots of noise dimensions in it. I tried using Is there a way to create arrays and datasets with unnamed dimensions? Thanks in advance. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/4079/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
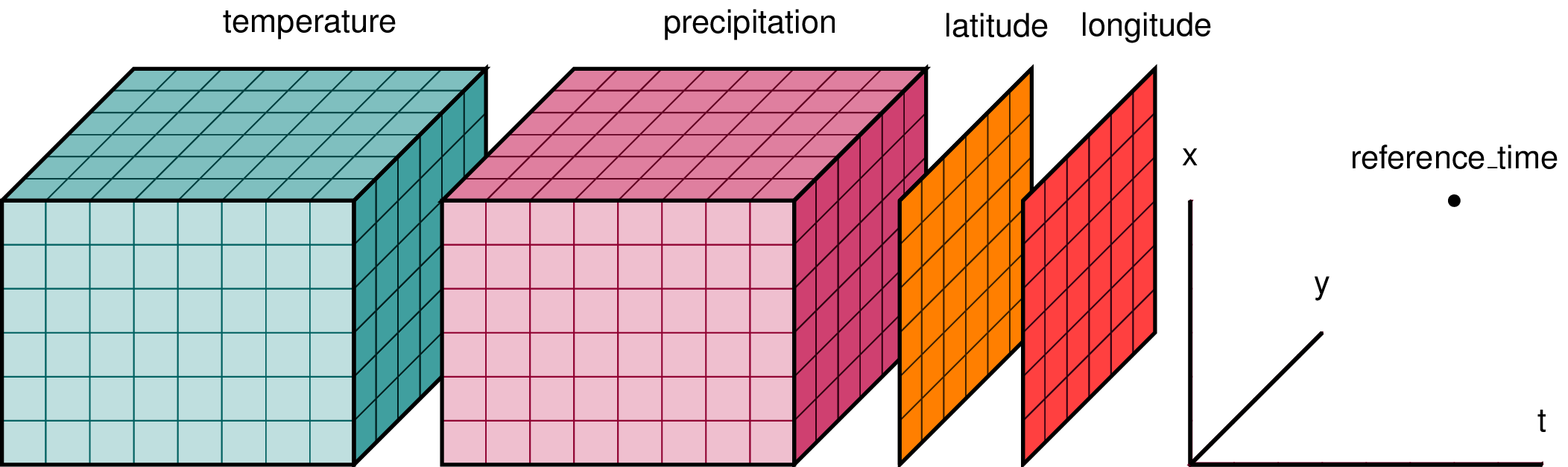
| 1300534066 | I_kwDOAMm_X85NhJMy | 6771 | Explaining xarray in a single picture | alimanfoo 703554 | open | 0 | 5 | 2022-07-11T10:47:55Z | 2022-07-20T09:41:07Z | CONTRIBUTOR | What is your issue?Hi folks, I'm working on a mini-tutorial introducing xarray for some folks in our genetics community and noticed something slightly confusing about the typical pictures used to help describe what xarray is for. E.g., this picture is commonly used:
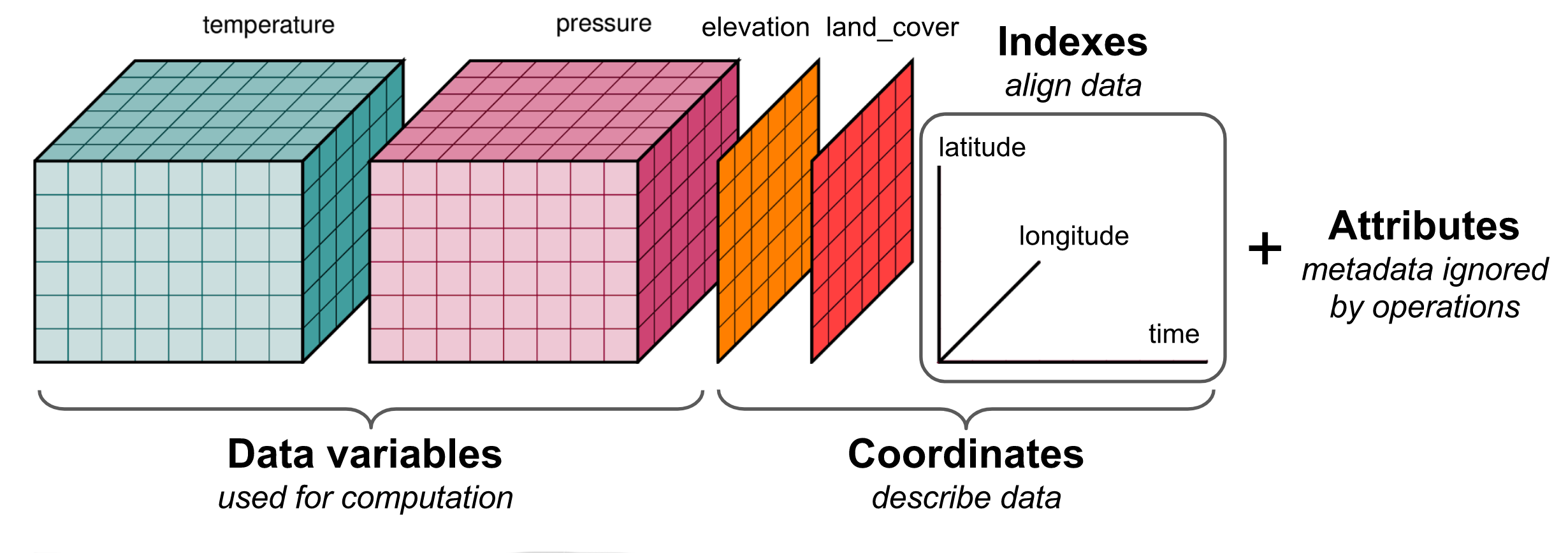
I get that temperature and precipitation are data variables which have been measured over the three dimensions of latitude, longitude and time. But I'm slightly confused here because I would've thought that latitude and longitude would be 1-dimensional coordinate variables, yet they are drawn as 2-D arrays? Elsewhere I found a slightly different version:
This makes more sense to me, because here the 2-D arrays have been labeled as "elevation" and "land_cover", and thus these are variables which are measured over the dimensions of latitude and longitude but not time, hence 2-D. Also, here latitude, longitude and time are shown labelling the dimensions, which again makes a bit more sense. However, "elevantion" and "land_cover" are included within the "coordinates" bracket, and I would have thought that elevation and land_cover would be more naturally considered as data variables? Feel free to close/ignore/set me straight if I'm missing something here, but just thought I would raise to say that I was looking for a simple picture to help me explain what xarray is all about for newcomers and found these existing pictures a little confusing. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/6771/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 834972299 | MDU6SXNzdWU4MzQ5NzIyOTk= | 5054 | Fancy indexing a Dataset with dask DataArray causes excessive memory usage | alimanfoo 703554 | closed | 0 | 3 | 2021-03-18T15:45:08Z | 2021-03-18T16:58:44Z | 2021-03-18T16:04:36Z | CONTRIBUTOR | I have a dataset comprising several variables. All variables are dask arrays (e.g., backed by zarr). I would like to use one of these variables, which is a 1d boolean array, to index the other variables along a large single dimension. The boolean indexing array is about ~40 million items long, with ~20 million true values. If I do this all via dask (i.e., not using xarray) then I can index one dask array with another dask array via fancy indexing. The indexing array is not loaded into memory or computed. If I need to know the shape and chunks of the resulting arrays I can call If I do this via There is a follow-on issue which is if I then want to run a computation over one of the indexed arrays, if the indexing was done via xarray then that leads to a further blow-up of multiple GB of memory usage, if using dask distributed cluster. I think the underlying issue here is that the indexing array is loaded into memory, and then gets copied multiple times when the dask graph is constructed. If using a distributed scheduler, further copies get made during scheduling of any subsequent computation. I made a notebook which illustrates the increased memory usage during https://colab.research.google.com/drive/1bn7Sj0An7TehwltWizU8j_l2OvPeoJyo?usp=sharing This is possibly the same underlying issue (and use case) as raised by @eric-czech in #4663, so feel free to close this if you think it's a duplicate. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/5054/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 819911891 | MDExOlB1bGxSZXF1ZXN0NTgyOTQyMjMy | 4984 | Adds Dataset.query() method, analogous to pandas DataFrame.query() | alimanfoo 703554 | closed | 0 | 11 | 2021-03-02T11:08:42Z | 2021-03-16T18:28:09Z | 2021-03-16T17:28:15Z | CONTRIBUTOR | 0 | pydata/xarray/pulls/4984 | This PR adds a
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/4984/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | pull | |||||
| 303270676 | MDU6SXNzdWUzMDMyNzA2NzY= | 1974 | xarray/zarr cloud demo | alimanfoo 703554 | closed | 0 | 15 | 2018-03-07T21:43:40Z | 2018-03-09T04:56:38Z | 2018-03-09T04:56:38Z | CONTRIBUTOR | Apologies for the very short notice, but I'm giving a webinar on zarr tomorrow and would like to include an xarray demo, especially if it included computing on some pangeo data stored in the cloud. Wouldn't have to be complex at all, just proof of concept. Does anyone have a code example I could borrow? cc @rabernat @jhamman @shoyer @mrocklin |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/1974/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 33396232 | MDExOlB1bGxSZXF1ZXN0MTU4MjA2NTI= | 127 | initial implementation of support for NetCDF groups | alimanfoo 703554 | closed | 0 | 0.1.1 664063 | 6 | 2014-05-13T13:12:53Z | 2014-06-27T17:23:33Z | 2014-05-16T01:46:09Z | CONTRIBUTOR | 0 | pydata/xarray/pulls/127 | Just to start getting familiar with xray, I've had a go at implementing support for opening a dataset from a specific group within a NetCDF file. I haven't tested on real data but there are a couple of unit tests covering simple cases. Let me know if you'd like to take this forward, happy to work on it further. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/127/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | pull |
Advanced export
JSON shape: default, array, newline-delimited, object
CREATE TABLE [issues] (
[id] INTEGER PRIMARY KEY,
[node_id] TEXT,
[number] INTEGER,
[title] TEXT,
[user] INTEGER REFERENCES [users]([id]),
[state] TEXT,
[locked] INTEGER,
[assignee] INTEGER REFERENCES [users]([id]),
[milestone] INTEGER REFERENCES [milestones]([id]),
[comments] INTEGER,
[created_at] TEXT,
[updated_at] TEXT,
[closed_at] TEXT,
[author_association] TEXT,
[active_lock_reason] TEXT,
[draft] INTEGER,
[pull_request] TEXT,
[body] TEXT,
[reactions] TEXT,
[performed_via_github_app] TEXT,
[state_reason] TEXT,
[repo] INTEGER REFERENCES [repos]([id]),
[type] TEXT
);
CREATE INDEX [idx_issues_repo]
ON [issues] ([repo]);
CREATE INDEX [idx_issues_milestone]
ON [issues] ([milestone]);
CREATE INDEX [idx_issues_assignee]
ON [issues] ([assignee]);
CREATE INDEX [idx_issues_user]
ON [issues] ([user]);