issues
23 rows where repo = 13221727, type = "issue" and user = 1200058 sorted by updated_at descending
This data as json, CSV (advanced)
Suggested facets: comments, state_reason, created_at (date), updated_at (date), closed_at (date)
| id | node_id | number | title | user | state | locked | assignee | milestone | comments | created_at | updated_at ▲ | closed_at | author_association | active_lock_reason | draft | pull_request | body | reactions | performed_via_github_app | state_reason | repo | type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 884649380 | MDU6SXNzdWU4ODQ2NDkzODA= | 5287 | Support for pandas Extension Arrays | Hoeze 1200058 | closed | 0 | 8 | 2021-05-10T17:00:17Z | 2024-04-18T12:52:04Z | 2024-04-18T12:52:04Z | NONE | Is your feature request related to a problem? Please describe.
I started writing an ExtensionArray which is basically a This is working great in Pandas, I can read and write Parquet as well as csv with it.
However, as soon as I'm using any Describe the solution you'd like Would it be possible to support Pandas Extension Types on coordinates? It's not necessary to compute anything on them, I'd just like to use them for dimensions. Describe alternatives you've considered I was thinking over implementing a NumPy duck array, but I have never tried this and it looks quite complicated compared to the Pandas Extension types. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/5287/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 384002323 | MDU6SXNzdWUzODQwMDIzMjM= | 2570 | np.clip() executes eagerly | Hoeze 1200058 | closed | 0 | 4 | 2018-11-24T16:25:03Z | 2023-12-03T05:29:17Z | 2023-12-03T05:29:17Z | NONE | Example:
Problem descriptionUsing np.clip() directly calculates the result, while xr.DataArray.clip() does not. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2570/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
not_planned | xarray 13221727 | issue | ||||||
| 595784008 | MDU6SXNzdWU1OTU3ODQwMDg= | 3945 | Implement `value_counts` method | Hoeze 1200058 | open | 0 | 3 | 2020-04-07T11:05:06Z | 2023-09-12T15:47:22Z | NONE | Implement MCVE Code Sample
Suggested API:
Expected Output
Problem DescriptionCurrently there is no existing equivalent to this method that I know in xarray. VersionsOutput of `xr.show_versions()`INSTALLED VERSIONS ------------------ commit: None python: 3.7.6 | packaged by conda-forge | (default, Jan 7 2020, 22:33:48) [GCC 7.3.0] python-bits: 64 OS: Linux OS-release: 5.3.11-1.el7.elrepo.x86_64 machine: x86_64 processor: byteorder: little LC_ALL: None LANG: en_US.UTF-8 LOCALE: en_US.UTF-8 libhdf5: 1.10.5 libnetcdf: 4.7.3 xarray: 0.15.0 pandas: 1.0.0 numpy: 1.17.5 scipy: 1.4.1 netCDF4: 1.5.3 pydap: None h5netcdf: 0.7.4 h5py: 2.10.0 Nio: None zarr: 2.4.0 cftime: 1.0.4.2 nc_time_axis: None PseudoNetCDF: None rasterio: None cfgrib: None iris: None bottleneck: None dask: 2.10.1 distributed: 2.10.0 matplotlib: 3.1.3 cartopy: None seaborn: 0.10.0 numbagg: None setuptools: 45.1.0.post20200119 pip: 20.0.2 conda: None pytest: 5.3.5 IPython: 7.12.0 sphinx: 2.0.1 |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3945/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 712217045 | MDU6SXNzdWU3MTIyMTcwNDU= | 4476 | Reimplement GroupBy.argmax | Hoeze 1200058 | open | 0 | 5 | 2020-09-30T19:25:22Z | 2023-03-03T06:59:40Z | NONE | Please implement Is your feature request related to a problem? Please describe.
Observed:
AttributeError Traceback (most recent call last) <ipython-input-84-15c199b0f7d4> in <module> ----> 1 da.groupby("g").argmax(dim="t") AttributeError: 'DataArrayGroupBy' object has no attribute 'argmax' ``` Describe the solution you'd like
Expected: Vector of length Workaround:
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/4476/reactions",
"total_count": 2,
"+1": 2,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 481838855 | MDU6SXNzdWU0ODE4Mzg4NTU= | 3224 | Add "on"-parameter to "merge" method | Hoeze 1200058 | closed | 0 | 2 | 2019-08-17T02:44:46Z | 2022-04-18T15:57:09Z | 2022-04-18T15:57:09Z | NONE | I'd like to propose a change to the merge method. Often, I meet cases where I'd like to merge subsets of the same dataset. However, this currently requires renaming of all dimensions, changing indices and merging them by hand. As an example, please consider the following dataset:
Now, I'd like to plot all values in To simplify this task, I'd like to have the following abstraction: ```python3 select tissuestissue_1 = ds.sel(observations = (ds.subtissue == "Whole_Blood")) tissue_2 = ds.sel(observations = (ds.subtissue == "Adipose_Subcutaneous")) inner join by individualmerged = tissue_1.merge(tissue_2, on="individual", newdim="merge_dim", join="inner") print(merged)
To summarize, I'd propose the following changes:
- Add parameter In case if the What do you think about this addition? |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3224/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 510844652 | MDU6SXNzdWU1MTA4NDQ2NTI= | 3432 | Scalar slice of MultiIndex is turned to tuples | Hoeze 1200058 | closed | 0 | 5 | 2019-10-22T18:55:52Z | 2022-03-17T17:11:41Z | 2022-03-17T17:11:41Z | NONE | Today I updated to I tried to select one observation of the following dataset:
As you can see, observations is now a tuple of Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3432/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
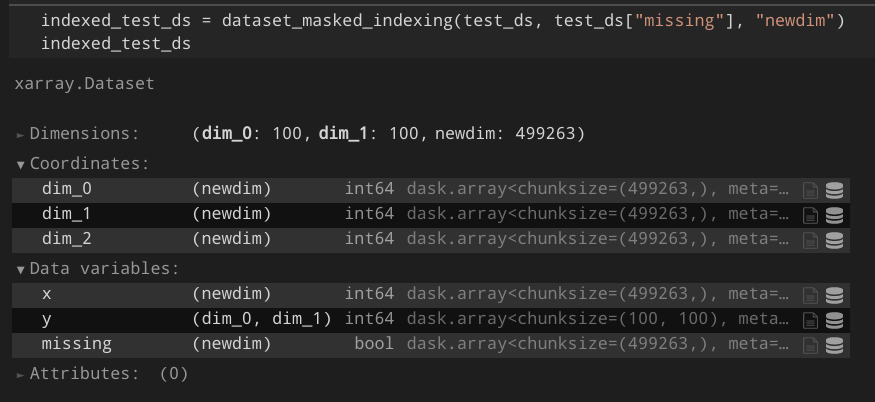
| 860418546 | MDU6SXNzdWU4NjA0MTg1NDY= | 5179 | N-dimensional boolean indexing | Hoeze 1200058 | open | 0 | 6 | 2021-04-17T14:07:48Z | 2021-07-16T17:30:45Z | NONE | Currently, the docs state that boolean indexing is only possible with 1-dimensional arrays: http://xarray.pydata.org/en/stable/indexing.html However, I often have the case where I'd like to convert a subset of an xarray to a dataframe.
Usually, I would call e.g.:
However, this approach is incredibly slow and memory-demanding, since it creates a MultiIndex of every possible coordinate in the array. Describe the solution you'd like
A better approach would be to directly allow index selection with the boolean array:
Additional context I created a proof-of-concept that works for my projects: https://gist.github.com/Hoeze/c746ea1e5fef40d99997f765c48d3c0d Some important lines are those: ```python def core_dim_locs_from_cond(cond, new_dim_name, core_dims=None) -> List[Tuple[str, xr.DataArray]]: [...] core_dim_locs = np.argwhere(cond.data) if isinstance(core_dim_locs, dask.array.core.Array): core_dim_locs = core_dim_locs.persist().compute_chunk_sizes() def subset_variable(variable, core_dim_locs, new_dim_name, mask=None): [...] subset = dask.array.asanyarray(variable.data)[mask] # force-set chunk size from known chunks chunk_sizes = core_dim_locs[0][1].chunks[0] subset._chunks = (chunk_sizes, *subset._chunks[1:]) ``` As a result, I would expect something like this:
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/5179/reactions",
"total_count": 2,
"+1": 2,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 489825483 | MDU6SXNzdWU0ODk4MjU0ODM= | 3281 | [proposal] concatenate by axis, ignore dimension names | Hoeze 1200058 | open | 0 | 4 | 2019-09-05T15:06:22Z | 2021-07-08T17:42:53Z | NONE | Hi, I wrote a helper function which allows to concatenate arrays like I often need this to combine very different feature types. ```python from typing import Union, Tuple, List import numpy as np import xarray as xr def concat_by_axis(
darrs: Union[List[xr.DataArray], Tuple[xr.DataArray]],
dims: Union[List[str], Tuple[str]],
axis: int = None,
**kwargs
):
"""
Concat arrays along some axis similar to ``` Would it make sense to include this in xarray? |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3281/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 636512559 | MDU6SXNzdWU2MzY1MTI1NTk= | 4143 | [Feature request] Masked operations | Hoeze 1200058 | open | 0 | 1 | 2020-06-10T20:04:45Z | 2021-04-22T20:54:03Z | NONE | Xarray already has Logically, a sparse array is equal to a masked dense array.
They only differ in their internal data representation.
Therefore, I would propose to have a This would solve a number of problems: - No more conversion of int -> float - Explicit value for missingness - When stacking data with missing values, the missing values can be just dropped - When converting data with missing values to DataFrame, the missing values can be just dropped MCVE Code SampleAn example would be outer joins with slightly different coordinates (taken from the documentation): ```python
Non-masked outer join:```python
The masked version:```python
Related issue: https://github.com/pydata/xarray/issues/3955 |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/4143/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 325475880 | MDU6SXNzdWUzMjU0NzU4ODA= | 2173 | Formatting error in conjunction with pandas.DataFrame | Hoeze 1200058 | closed | 0 | 6 | 2018-05-22T21:49:24Z | 2021-04-13T15:04:51Z | 2021-04-13T15:04:51Z | NONE | Code Sample, a copy-pastable example if possible```python import pandas as pd import numpy as np import xarray as xr sample_data = np.random.uniform(size=[2,2000,10000]) x = xr.Dataset({"sample_data": (sample_data.shape, sample_data)}) print(x) df = pd.DataFrame({"x": [1,2,3], "y": [2,4,6]})
x["df"] = df
print(x)
```
Problem descriptionPrinting a xarray.Dataset results in an error when containing a pandas.DataFrame Expected OutputShould print string representation of Dataset Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2173/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 512879550 | MDU6SXNzdWU1MTI4Nzk1NTA= | 3452 | [feature request] __iter__() for rolling-window on datasets | Hoeze 1200058 | open | 0 | 2 | 2019-10-26T20:08:06Z | 2021-02-18T21:41:58Z | NONE | Currently, rolling() on a dataset does not return an iterator: MCVE Code Sample```python arr = xr.DataArray(np.arange(0, 7.5, 0.5).reshape(3, 5), dims=('x', 'y')) r = arr.to_dataset(name="test").rolling(y=3)
for label, arr_window in r:
print(label)
TypeError Traceback (most recent call last) <ipython-input-12-b1703cb71c1e> in <module> 3 4 r = arr.to_dataset(name="test").rolling(y=3) ----> 5 for label, arr_window in r: 6 print(label) TypeError: 'DatasetRolling' object is not iterable ``` Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3452/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 528060435 | MDU6SXNzdWU1MjgwNjA0MzU= | 3570 | fillna on dataset converts all variables to float | Hoeze 1200058 | open | 0 | 5 | 2019-11-25T12:39:49Z | 2020-09-15T15:35:04Z | NONE | MCVE Code Sample
Expected Output
Problem DescriptionI'd like to use Would it be possible to apply Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3570/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 566509807 | MDU6SXNzdWU1NjY1MDk4MDc= | 3775 | [Question] Efficient shortcut for unstacking only parts of dimension? | Hoeze 1200058 | open | 0 | 1 | 2020-02-17T20:46:03Z | 2020-03-07T04:53:05Z | NONE | Hi all, is there an efficient way to unstack only parts of a MultiIndex? Consider for example the following array:
Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3775/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 511498714 | MDU6SXNzdWU1MTE0OTg3MTQ= | 3438 | Re-indexing causes coordinates to be dropped | Hoeze 1200058 | closed | 0 | 2 | 2019-10-23T18:31:18Z | 2020-01-09T01:46:46Z | 2020-01-09T01:46:46Z | NONE | Hi, I encounter a problem with the index being dropped when I rename a dimension and stack it afterwards: MCVE Code Sample```python ds = xr.Dataset({ "test": xr.DataArray( [[[1,2],[3,4]], [[1,2],[3,4]]], dims=("genes", "observations", "subtissues"), coords={ "observations": xr.DataArray(["x-1", "y-1"], dims=("observations",)), "individuals": xr.DataArray(["x", "y"], dims=("observations",)), "genes": xr.DataArray(["a", "b"], dims=("genes",)), "subtissues": xr.DataArray(["c", "d"], dims=("subtissues",)), } ) })
Is this by intention? Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3438/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 522238536 | MDU6SXNzdWU1MjIyMzg1MzY= | 3518 | Have "unstack" return a boolean mask? | Hoeze 1200058 | closed | 0 | 1 | 2019-11-13T13:54:49Z | 2019-11-16T14:36:43Z | 2019-11-16T14:36:43Z | NONE | MCVE Code Sample
Expected Output```python
Problem DescriptionUnstacking changes the data type to float for Currently, I obtain a boolean Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3518/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 491172429 | MDU6SXNzdWU0OTExNzI0Mjk= | 3296 | [Docs] parameters + data type broken | Hoeze 1200058 | closed | 0 | 2 | 2019-09-09T15:36:40Z | 2019-09-09T15:41:36Z | 2019-09-09T15:40:29Z | NONE | Hi, since this is already present some time and I could not find a corresponding issue: The documentation format seems to be broken. Parameter name and data type stick together:
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3296/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 325661581 | MDU6SXNzdWUzMjU2NjE1ODE= | 2175 | [Feature Request] Visualizing dimensions | Hoeze 1200058 | open | 0 | 4 | 2018-05-23T11:22:29Z | 2019-07-12T16:10:23Z | NONE | Hi, I'm curious how you created your logo:
I'd like to create visualizations of the dimensions in my dataset similar to your logo. Having a functionality simplifying this task would be a very useful feature in xarray. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2175/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
xarray 13221727 | issue | ||||||||
| 338226520 | MDU6SXNzdWUzMzgyMjY1MjA= | 2267 | Some simple broadcast_dim method? | Hoeze 1200058 | closed | 0 | 9 | 2018-07-04T10:48:27Z | 2019-07-06T13:06:45Z | 2019-07-06T13:06:45Z | NONE | I've already found xr.broadcast(arrays). However, I'd like to just add a new dimension with a specific size to one DataArray. I could not find any simple option to do this. If there is no such option: - add a size parameter to DataArray.expand_dims? - DataArray.broadcast_dims({"a": M, "b": N})? |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2267/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 337619718 | MDU6SXNzdWUzMzc2MTk3MTg= | 2263 | [bug] Exception ignored in generator object Variable | Hoeze 1200058 | closed | 0 | 9 | 2018-07-02T18:30:57Z | 2019-01-23T00:56:19Z | 2019-01-23T00:56:18Z | NONE |
Problem descriptionDuring this, the following warning pops up: ``` Exception ignored in: <generator object Variable._broadcast_indexes.\<locals>.\<genexpr> at 0x7fcdd479f1a8> Traceback (most recent call last): File "/usr/local/lib/python3.6/dist-packages/xarray/core/variable.py", line 470, in <genexpr> if all(isinstance(k, BASIC_INDEXING_TYPES) for k in key): SystemError: error return without exception set ``` Expected OutputNo error Possible solution:Each time I execute: Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2263/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 336220647 | MDU6SXNzdWUzMzYyMjA2NDc= | 2253 | autoclose=True is not implemented for the h5netcdf backend | Hoeze 1200058 | closed | 0 | 2 | 2018-06-27T13:03:44Z | 2019-01-13T01:38:24Z | 2019-01-13T01:38:24Z | NONE | Hi, are there any plans to enable Error message:
Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2253/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 378326194 | MDU6SXNzdWUzNzgzMjYxOTQ= | 2549 | to_dask_dataframe for xr.DataArray | Hoeze 1200058 | closed | 0 | 2 | 2018-11-07T15:02:22Z | 2018-11-07T16:27:56Z | 2018-11-07T16:27:56Z | NONE | Is there some xr.DataArray.to_dask_dataframe() method? |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2549/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 325470877 | MDU6SXNzdWUzMjU0NzA4Nzc= | 2172 | Errors on pycharm completion | Hoeze 1200058 | closed | 0 | 2 | 2018-05-22T21:31:42Z | 2018-05-27T20:48:30Z | 2018-05-27T20:48:30Z | NONE | Code Sample, a copy-pastable example if possible```python execute:import numpy as np import xarray as xr sample_data = np.random.uniform(size=[2,2000,10000]) x = xr.Dataset({"sample_data": (sample_data.shape, sample_data)}) x2 = x["sample_data"] now type by hand:x2. ```
Problem descriptionI'm not completely sure if it's a xarray problem, but each time I enter [some dataset]. (note the point) inside PyCharm's python console, I get a python exception instead of some autocompletion. Expected Output
Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2172/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 325889600 | MDU6SXNzdWUzMjU4ODk2MDA= | 2177 | Dataset.to_netcdf() cannot create group with engine="h5netcdf" | Hoeze 1200058 | closed | 0 | 1 | 2018-05-23T22:03:07Z | 2018-05-25T00:52:07Z | 2018-05-25T00:52:07Z | NONE | Code Sample, a copy-pastable example if possible```python import pandas as pd import numpy as np import xarray as xr sample_data = np.random.uniform(size=[2,2000,10000]) x = xr.Dataset({"sample_data": (("x", "y", "z"), sample_data)}) df = pd.DataFrame({"x": [1,2,3], "y": [2,4,6]}) x["df"] = df print(x) not working:x.to_netcdf("test.h5", group="asdf", engine="h5netcdf") working:x.to_netcdf("test.h5", group="asdf", engine="netcdf4") ```
Problem descriptionh5netcdf does not allow creating groups Expected Outputshould save data to "test.h5" Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2177/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue |
Advanced export
JSON shape: default, array, newline-delimited, object
CREATE TABLE [issues] (
[id] INTEGER PRIMARY KEY,
[node_id] TEXT,
[number] INTEGER,
[title] TEXT,
[user] INTEGER REFERENCES [users]([id]),
[state] TEXT,
[locked] INTEGER,
[assignee] INTEGER REFERENCES [users]([id]),
[milestone] INTEGER REFERENCES [milestones]([id]),
[comments] INTEGER,
[created_at] TEXT,
[updated_at] TEXT,
[closed_at] TEXT,
[author_association] TEXT,
[active_lock_reason] TEXT,
[draft] INTEGER,
[pull_request] TEXT,
[body] TEXT,
[reactions] TEXT,
[performed_via_github_app] TEXT,
[state_reason] TEXT,
[repo] INTEGER REFERENCES [repos]([id]),
[type] TEXT
);
CREATE INDEX [idx_issues_repo]
ON [issues] ([repo]);
CREATE INDEX [idx_issues_milestone]
ON [issues] ([milestone]);
CREATE INDEX [idx_issues_assignee]
ON [issues] ([assignee]);
CREATE INDEX [idx_issues_user]
ON [issues] ([user]);

 Source:
Source: 


