issue_comments
13 rows where user = 12760310 sorted by updated_at descending
This data as json, CSV (advanced)
Suggested facets: issue_url, created_at (date), updated_at (date)
user 1
- guidocioni · 13 ✖
| id | html_url | issue_url | node_id | user | created_at | updated_at ▲ | author_association | body | reactions | performed_via_github_app | issue |
|---|---|---|---|---|---|---|---|---|---|---|---|
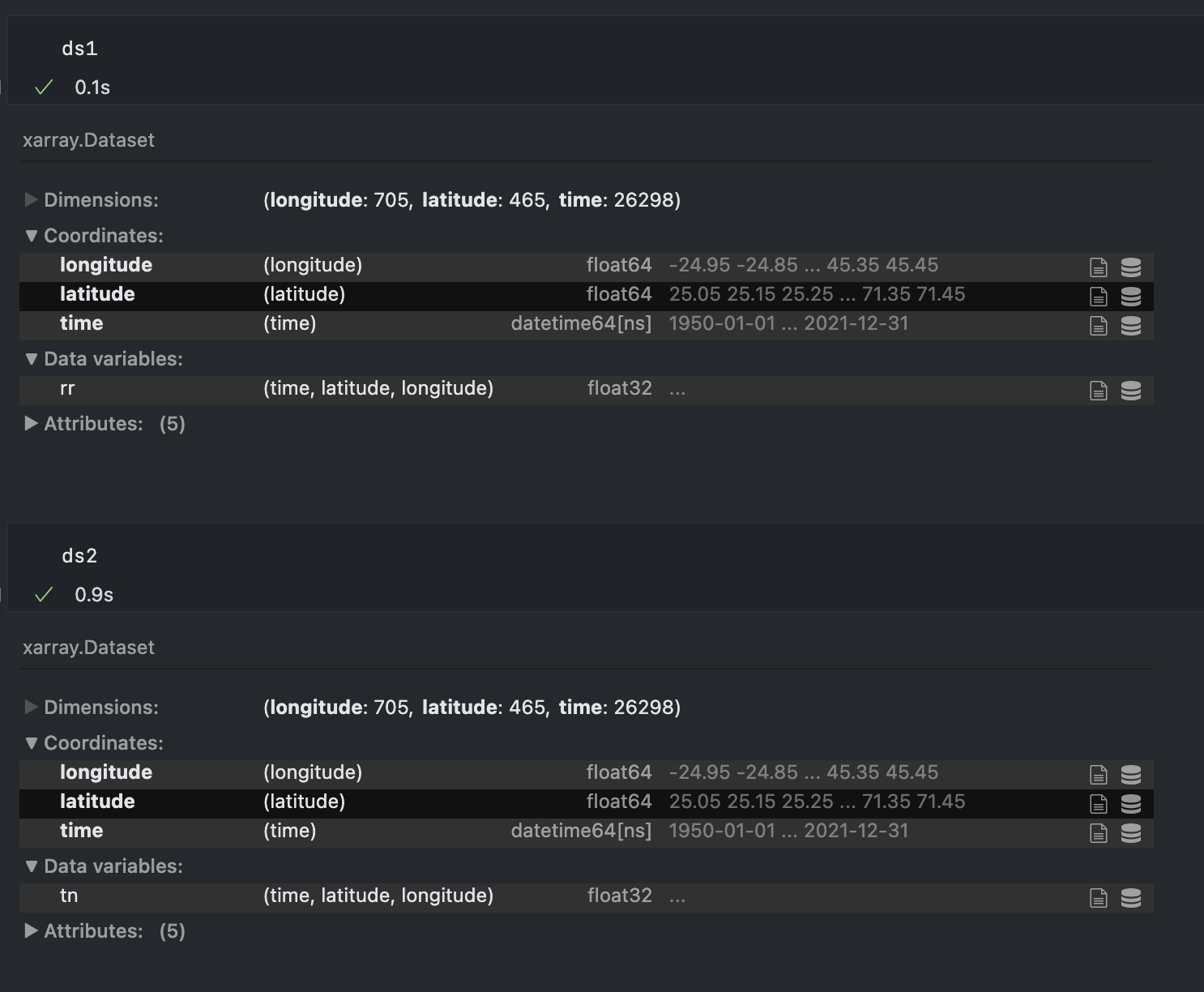
| 1260899163 | https://github.com/pydata/xarray/issues/7065#issuecomment-1260899163 | https://api.github.com/repos/pydata/xarray/issues/7065 | IC_kwDOAMm_X85LJ8tb | guidocioni 12760310 | 2022-09-28T13:16:13Z | 2022-09-28T13:16:13Z | NONE | Hey @benbovy, sorry for resurrect again this post but today I'm seeing the same issue and for the love of me I cannot understand what is the difference in this dataset that is causing the latitude and longitude arrays to be duplicated...
If I try to merge these two datasets I get one with lat lon doubled in size. |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
Merge wrongfully creating NaN 1381955373 | |
| 1255092548 | https://github.com/pydata/xarray/issues/7065#issuecomment-1255092548 | https://api.github.com/repos/pydata/xarray/issues/7065 | IC_kwDOAMm_X85KzzFE | guidocioni 12760310 | 2022-09-22T14:17:07Z | 2022-09-22T14:17:17Z | NONE |
The differences are larger than I would expect (order of 0.1 in some variables) but could be related to the fact that, when using different precisions, the closest grid points to the target point could change. This would eventually lead to a different value of the variable extracted from the original dataset. Unfortunately I didn't have time to verify if it was the case, but I think this is the only valid explanation because the variables of the dataset are untouched. It is still puzzling because, as the target points have a precision of e.g. (45.820497820, 13.003510004), I would expect the cast of the dataset coordinates from e.g. (45.8, 13.0) to preserve the 0 (45.800000000, 13.00000000), so that the closest point should not change. Anyway, I think we're getting off-topic, thanks for the help :) |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
Merge wrongfully creating NaN 1381955373 | |
| 1255026304 | https://github.com/pydata/xarray/issues/7065#issuecomment-1255026304 | https://api.github.com/repos/pydata/xarray/issues/7065 | IC_kwDOAMm_X85Kzi6A | guidocioni 12760310 | 2022-09-22T13:28:17Z | 2022-09-22T13:28:31Z | NONE | Mmmm that's weird, because the execution time is really different, and it would be hard to explain it if all the arrays are casted to the same Yeah, for the nearest lookup I already implemented "my version" of |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
Merge wrongfully creating NaN 1381955373 | |
| 1254985357 | https://github.com/pydata/xarray/issues/7065#issuecomment-1254985357 | https://api.github.com/repos/pydata/xarray/issues/7065 | IC_kwDOAMm_X85KzY6N | guidocioni 12760310 | 2022-09-22T12:56:35Z | 2022-09-22T12:56:35Z | NONE | Sorry, that brings me to another question that I never even considered. As my latitude and longitude arrays in both datasets have a resolution of 0.1 degrees, wouldn't it make sense to use From this dataset I'm extracting the closest points to a station inside a user-defined radius, doing something similar to
The thing is, if I use |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
Merge wrongfully creating NaN 1381955373 | |
| 1254941693 | https://github.com/pydata/xarray/issues/7065#issuecomment-1254941693 | https://api.github.com/repos/pydata/xarray/issues/7065 | IC_kwDOAMm_X85KzOP9 | guidocioni 12760310 | 2022-09-22T12:17:10Z | 2022-09-22T12:17:10Z | NONE | @benbovy you have no idea how much time I spent trying to understand what the difference between the two different datasets was....and I completely missed the The problem is that I tried to merge with Before closing, just a curiosity: in this corner case shouldn't |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
Merge wrongfully creating NaN 1381955373 | |
| 1210383450 | https://github.com/pydata/xarray/issues/6904#issuecomment-1210383450 | https://api.github.com/repos/pydata/xarray/issues/6904 | IC_kwDOAMm_X85IJPxa | guidocioni 12760310 | 2022-08-10T09:07:00Z | 2022-08-10T09:07:00Z | NONE | This is a minimal working example that I could come up with. You can try to open any netcdf that you have. I tested on a small one and it didn't reproduce the error, so it is definitely only happening with large datasets when the arrays are not loaded into memory. Unfortunately, as you need a large file, I cannot really attach it here. ```python import xarray as xr from tqdm.contrib.concurrent import process_map import pprint def main(): global ds ds = xr.open_dataset('input.nc') it = range(0, 5) results = [] for i in it: results.append(compute(i)) print("------------Serial results-----------------") pprint.pprint(results) results = process_map(compute, it, max_workers=6, chunksize=1, disable=True) print("------------Parallel results-----------------") pprint.pprint(results) def compute(station): ds_point = ds.isel(lat=0, lon=0) return station, ds_point.t_2m_max.mean().item(), ds_point.t_2m_min.mean().item(), ds_point.lon.min().item(), ds_point.lat.min().item() if name == "main": main() ``` |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
`sel` behaving randomly when applying to a dataset with multiprocessing 1333650265 | |
| 1210349031 | https://github.com/pydata/xarray/issues/6904#issuecomment-1210349031 | https://api.github.com/repos/pydata/xarray/issues/6904 | IC_kwDOAMm_X85IJHXn | guidocioni 12760310 | 2022-08-10T08:38:31Z | 2022-08-10T08:38:31Z | NONE |
Ok, it seems to fail also with exact lookups o.O This is extremely weird I'm using
Result for the serial version
And for the parallel version with EXACTLY the same code
|
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
`sel` behaving randomly when applying to a dataset with multiprocessing 1333650265 | |
| 1210341456 | https://github.com/pydata/xarray/issues/6904#issuecomment-1210341456 | https://api.github.com/repos/pydata/xarray/issues/6904 | IC_kwDOAMm_X85IJFhQ | guidocioni 12760310 | 2022-08-10T08:32:13Z | 2022-08-10T08:32:13Z | NONE |
That causes an error
Here is the complete tracebabk
I think we may be heading the right direction |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
`sel` behaving randomly when applying to a dataset with multiprocessing 1333650265 | |
| 1210285626 | https://github.com/pydata/xarray/issues/6904#issuecomment-1210285626 | https://api.github.com/repos/pydata/xarray/issues/6904 | IC_kwDOAMm_X85II346 | guidocioni 12760310 | 2022-08-10T07:41:20Z | 2022-08-10T07:41:20Z | NONE |
mmm ok I'll try and let you know. BTW is there any advantage or difference in terms of cpu and memory consumption in opening the file only one or let it open by every process? I'm asking because I thought opening in every process was just plain stupid but it seems to perform exactly the same, so maybe I'm just creating a problem where there is none |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
`sel` behaving randomly when applying to a dataset with multiprocessing 1333650265 | |
| 1210238864 | https://github.com/pydata/xarray/issues/6904#issuecomment-1210238864 | https://api.github.com/repos/pydata/xarray/issues/6904 | IC_kwDOAMm_X85IIseQ | guidocioni 12760310 | 2022-08-10T06:51:18Z | 2022-08-10T06:51:18Z | NONE |
ok that's a good shot.
Will that work in the same way if I still use |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
`sel` behaving randomly when applying to a dataset with multiprocessing 1333650265 | |
| 1210220238 | https://github.com/pydata/xarray/issues/6904#issuecomment-1210220238 | https://api.github.com/repos/pydata/xarray/issues/6904 | IC_kwDOAMm_X85IIn7O | guidocioni 12760310 | 2022-08-10T06:30:06Z | 2022-08-10T06:30:06Z | NONE |
I haven't tried yet because it doesn't really match my use case. One idea that I had was to provide the list of points before starting the loop, creating an iterator with the slices from the xarray and then pass this to the loop. But I would end up using more data than necessary because I don't process all cases. another thing that I've noticed is that if the list of iterators is smaller than the chunksize everything's good, probably because it reverts to the serial case as only 1 worker is processing |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
`sel` behaving randomly when applying to a dataset with multiprocessing 1333650265 | |
| 1210174583 | https://github.com/pydata/xarray/issues/6904#issuecomment-1210174583 | https://api.github.com/repos/pydata/xarray/issues/6904 | IC_kwDOAMm_X85IIcx3 | guidocioni 12760310 | 2022-08-10T05:23:13Z | 2022-08-10T05:24:24Z | NONE |
Yep, and yep (believe me, I've tried anything in desperation 😄)
Which method should I use then? I need the closest point
Yep I can try to make a minimal example, however, in order to reproduce the issue, I think it's necessary to open a large dataset. |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
`sel` behaving randomly when applying to a dataset with multiprocessing 1333650265 | |
| 1206361835 | https://github.com/pydata/xarray/issues/6879#issuecomment-1206361835 | https://api.github.com/repos/pydata/xarray/issues/6879 | IC_kwDOAMm_X85H557r | guidocioni 12760310 | 2022-08-05T11:53:15Z | 2022-08-05T11:53:30Z | NONE |
Hey, thanks for the workaround.
However, I'm still not convinced that this is the "correct" behaviour.
If What is the use case in enlarging the 1-D array to a 3-D array with coordinates that it didn't have before? |
{
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
`Dataset.where()` incorrectly applies mask and creates new dimensions 1329754426 |
Advanced export
JSON shape: default, array, newline-delimited, object
CREATE TABLE [issue_comments] (
[html_url] TEXT,
[issue_url] TEXT,
[id] INTEGER PRIMARY KEY,
[node_id] TEXT,
[user] INTEGER REFERENCES [users]([id]),
[created_at] TEXT,
[updated_at] TEXT,
[author_association] TEXT,
[body] TEXT,
[reactions] TEXT,
[performed_via_github_app] TEXT,
[issue] INTEGER REFERENCES [issues]([id])
);
CREATE INDEX [idx_issue_comments_issue]
ON [issue_comments] ([issue]);
CREATE INDEX [idx_issue_comments_user]
ON [issue_comments] ([user]);
issue 3