issues
3 rows where user = 6249613 sorted by updated_at descending
This data as json, CSV (advanced)
Suggested facets: comments, created_at (date), updated_at (date), closed_at (date)
| id | node_id | number | title | user | state | locked | assignee | milestone | comments | created_at | updated_at ▲ | closed_at | author_association | active_lock_reason | draft | pull_request | body | reactions | performed_via_github_app | state_reason | repo | type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1863728367 | I_kwDOAMm_X85vFjzv | 8106 | Plotting accessor error for datasets with Dimension name matches coordinate names | blaylockbk 6249613 | closed | 0 | 3 | 2023-08-23T17:20:09Z | 2023-09-22T12:48:35Z | 2023-09-22T12:48:35Z | NONE | What happened?I am working with NetCDF files produced from the Model Evaluation Tools (MET). This feature in xarray v2023.8.0
lets me read the file where the dimension names ```python import urllib import xarray as xr download a sample MET fileurllib.request.urlretrieve( "https://github.com/dtcenter/MET/raw/main_v11.1/data/poly/NCEP_masks/WEST_mask.nc", "WEST_mask.nc", ) open MET fileds = xr.open_dataset(
"WEST_mask.nc",
engine="netcdf4",
)
ds
```
However, one issue I've run into is that the plotting accessor doesn't seem to work for this case ```python ds.WEST.plot() OUT: ValueError: coordinate 'lat' is a DataArray dimension, but it has shape (110, 147) rather than expected shape 110 matching the dimension size ``` To overcome this, I had to rename the dims
What did you expect to happen?I expected the plot accessor to work without needing to rename the dimensions. Minimal Complete Verifiable Example```Python import urllib import xarray as xr download a sample MET fileurllib.request.urlretrieve( "https://github.com/dtcenter/MET/raw/main_v11.1/data/poly/NCEP_masks/WEST_mask.nc", "WEST_mask.nc", ) ds = xr.open_dataset( "WEST_mask.nc", engine="netcdf4", ) ds.WEST.plot() ``` MVCE confirmation
Relevant log outputNo response Anything else we need to know?No response Environment
INSTALLED VERSIONS
------------------
commit: None
python: 3.11.4 | packaged by conda-forge | (main, Jun 10 2023, 18:08:17) [GCC 12.2.0]
python-bits: 64
OS: Linux
OS-release: 3.10.0-1160.53.1.el7.x86_64
machine: x86_64
processor: x86_64
byteorder: little
LC_ALL: None
LANG: en_US.UTF-8
LOCALE: ('en_US', 'UTF-8')
libhdf5: 1.12.2
libnetcdf: 4.9.1
xarray: 2023.8.0
pandas: 2.0.3
numpy: 1.25.2
scipy: 1.11.2
netCDF4: 1.6.3
pydap: None
h5netcdf: 1.2.0
h5py: 3.8.0
Nio: None
zarr: None
cftime: 1.6.2
nc_time_axis: None
PseudoNetCDF: None
iris: None
bottleneck: None
dask: 2023.8.1
distributed: 2023.8.1
matplotlib: 3.7.2
cartopy: 0.22.0
seaborn: 0.12.2
numbagg: None
fsspec: 2023.6.0
cupy: None
pint: 0.22
sparse: None
flox: None
numpy_groupies: None
setuptools: 68.1.2
pip: 23.2.1
conda: None
pytest: 7.4.0
mypy: None
IPython: 8.7.0
sphinx: 4.5.0
/p/home/blaylock/miniconda3/envs/flight/lib/python3.11/site-packages/_distutils_hack/__init__.py:33: UserWarning: Setuptools is replacing distutils.
warnings.warn("Setuptools is replacing distutils.")
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/8106/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
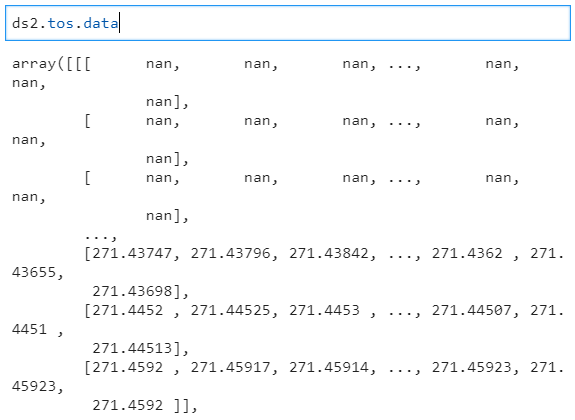
| 706507153 | MDU6SXNzdWU3MDY1MDcxNTM= | 4449 | Did copy(deep=True) break with 0.16.1? | blaylockbk 6249613 | closed | 0 | 4 | 2020-09-22T15:59:41Z | 2023-03-12T21:08:42Z | 2023-03-12T21:08:42Z | NONE | What happened: I have a script that downloads a file, reads and copies it to memory with What you expected to happen: In 0.16.0 and earlier, the variable data is available ( Minimal Complete Verifiable Example: ```python import xarray as xr import os import urllib.request Get sample NetCDF fileurl = 'https://www.unidata.ucar.edu/software/netcdf/examples/tos_O1_2001-2002.nc' FILE = 'tos_O1_2001-2002.nc' urllib.request.urlretrieve(url, FILE) Open the NetCDF fileds1 = xr.open_dataset(FILE) Make a copy of the Datasetds2 = ds1.copy(deep=True) and close the originalds1.close() remove the NetCDF fileos.remove(FILE) Read the copied datasetds2 ``` Anything else we need to know?:
Output for xarray v0.16.0
Output for xarray v0.16.1
Environment: Output of <tt>xr.show_versions()</tt> for xarray 0.16.0INSTALLED VERSIONS ------------------ commit: None python: 3.8.5 | packaged by conda-forge | (default, Sep 16 2020, 17:19:16) [MSC v.1916 64 bit (AMD64)] python-bits: 64 OS: Windows OS-release: 10 machine: AMD64 processor: Intel64 Family 6 Model 142 Stepping 12, GenuineIntel byteorder: little LC_ALL: None LANG: None LOCALE: English_United States.1252 libhdf5: 1.10.6 libnetcdf: 4.7.4 xarray: 0.16.0 pandas: 1.1.2 numpy: 1.19.1 scipy: 1.5.0 netCDF4: 1.5.4 pydap: None h5netcdf: None h5py: 2.10.0 Nio: None zarr: None cftime: 1.2.1 nc_time_axis: None PseudoNetCDF: None rasterio: None cfgrib: 0.9.8.4 iris: None bottleneck: None dask: None distributed: None matplotlib: 3.3.2 cartopy: 0.18.0 seaborn: None numbagg: None pint: 0.16 setuptools: 49.6.0.post20200917 pip: 20.2.3 conda: None pytest: None IPython: 7.18.1 sphinx: NoneOutput of <tt>xr.show_versions()</tt> for xarray 0.16.1INSTALLED VERSIONS ------------------ commit: None python: 3.8.5 | packaged by conda-forge | (default, Sep 16 2020, 17:19:16) [MSC v.1916 64 bit (AMD64)] python-bits: 64 OS: Windows OS-release: 10 machine: AMD64 processor: Intel64 Family 6 Model 142 Stepping 12, GenuineIntel byteorder: little LC_ALL: None LANG: None LOCALE: English_United States.1252 libhdf5: 1.10.6 libnetcdf: 4.7.4 xarray: 0.16.1 pandas: 1.1.2 numpy: 1.19.1 scipy: 1.5.0 netCDF4: 1.5.4 pydap: None h5netcdf: None h5py: 2.10.0 Nio: None zarr: None cftime: 1.2.1 nc_time_axis: None PseudoNetCDF: None rasterio: None cfgrib: 0.9.8.4 iris: None bottleneck: None dask: None distributed: None matplotlib: 3.3.2 cartopy: 0.18.0 seaborn: None numbagg: None pint: 0.16 setuptools: 49.6.0.post20200917 pip: 20.2.3 conda: None pytest: None IPython: 7.18.1 sphinx: Nonexarray: 0.16.0 pandas: 1.1.2 numpy: 1.19.1 scipy: 1.5.0 netCDF4: 1.5.4 pydap: None h5netcdf: None h5py: 2.10.0 Nio: None zarr: None cftime: 1.2.1 nc_time_axis: None PseudoNetCDF: None rasterio: None cfgrib: 0.9.8.4 iris: None bottleneck: None dask: None distributed: None matplotlib: 3.3.2 cartopy: 0.18.0 seaborn: None numbagg: None pint: 0.16 setuptools: 49.6.0.post20200917 pip: 20.2.3 conda: None pytest: None IPython: 7.18.1 sphinx: None |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/4449/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue | ||||||
| 588526556 | MDU6SXNzdWU1ODg1MjY1NTY= | 3898 | html repr does not work in Edge browser | blaylockbk 6249613 | closed | 0 | 4 | 2020-03-26T15:52:14Z | 2022-05-02T20:27:27Z | 2022-05-02T20:27:27Z | NONE | Just updated a conda environment and it installed xarray to 0.15.1. I was a bit lost with the new html output because it does not work when Jupyter is running in an Edge browser (not the chromium version). After I switched to Chrome I figured out that the html output was clickable and expanded the data I needed to see. Display in the Edge browser: sections do not expand when clicked
Display in the Chrome browser: sections expand when clicked
The text output is so engrained in my programming muscle memory, but I want to try this new html output...Can you add some documentation describing how to use the html output, especially details like browser requirements, what the display should look like, etc. |
{
"url": "https://api.github.com/repos/pydata/xarray/issues/3898/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | xarray 13221727 | issue |
Advanced export
JSON shape: default, array, newline-delimited, object
CREATE TABLE [issues] (
[id] INTEGER PRIMARY KEY,
[node_id] TEXT,
[number] INTEGER,
[title] TEXT,
[user] INTEGER REFERENCES [users]([id]),
[state] TEXT,
[locked] INTEGER,
[assignee] INTEGER REFERENCES [users]([id]),
[milestone] INTEGER REFERENCES [milestones]([id]),
[comments] INTEGER,
[created_at] TEXT,
[updated_at] TEXT,
[closed_at] TEXT,
[author_association] TEXT,
[active_lock_reason] TEXT,
[draft] INTEGER,
[pull_request] TEXT,
[body] TEXT,
[reactions] TEXT,
[performed_via_github_app] TEXT,
[state_reason] TEXT,
[repo] INTEGER REFERENCES [repos]([id]),
[type] TEXT
);
CREATE INDEX [idx_issues_repo]
ON [issues] ([repo]);
CREATE INDEX [idx_issues_milestone]
ON [issues] ([milestone]);
CREATE INDEX [idx_issues_assignee]
ON [issues] ([assignee]);
CREATE INDEX [idx_issues_user]
ON [issues] ([user]);