issues: 344621749
This data as json
| id | node_id | number | title | user | state | locked | assignee | milestone | comments | created_at | updated_at | closed_at | author_association | active_lock_reason | draft | pull_request | body | reactions | performed_via_github_app | state_reason | repo | type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
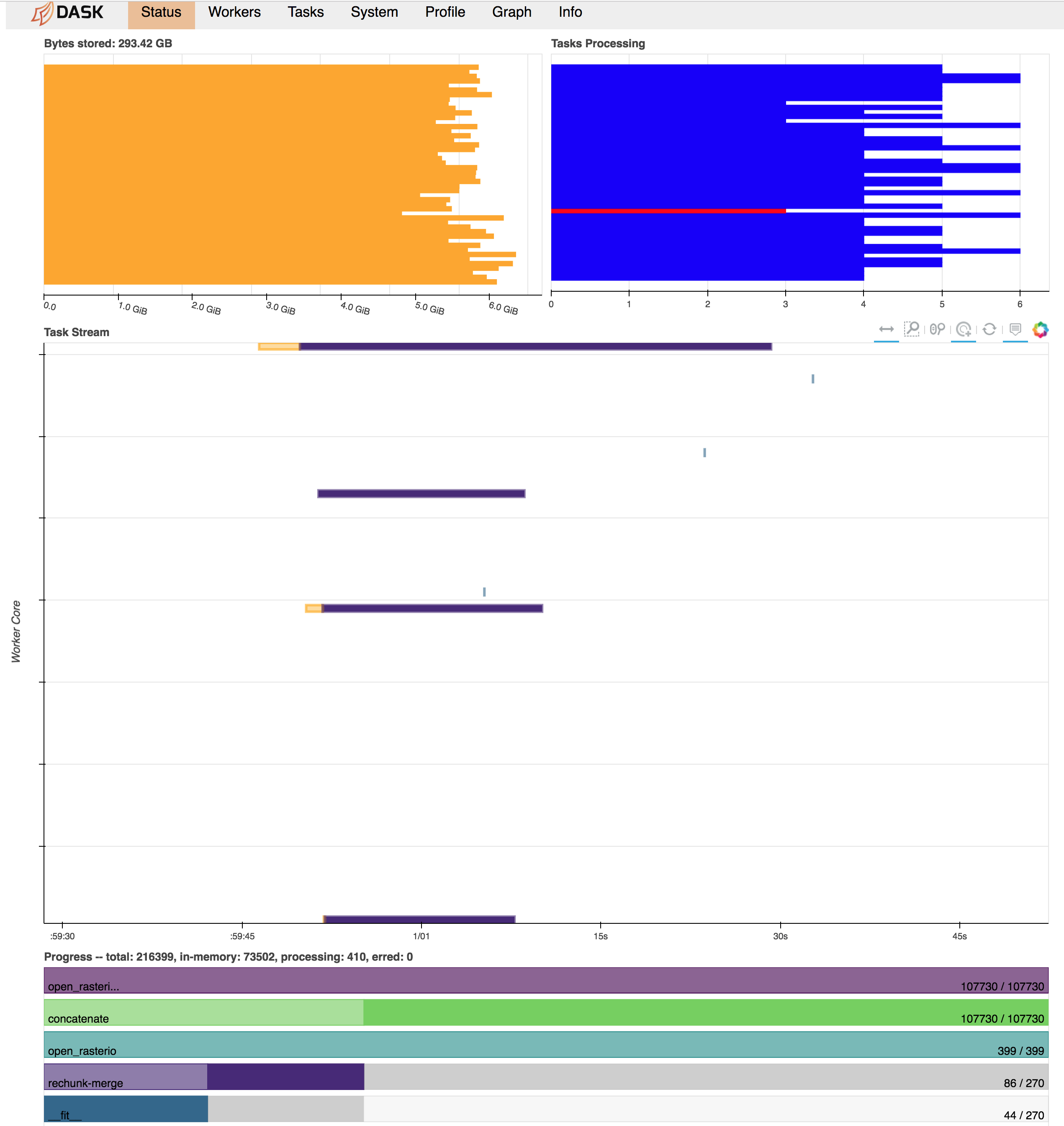
| 344621749 | MDU6SXNzdWUzNDQ2MjE3NDk= | 2314 | Chunked processing across multiple raster (geoTIF) files | 2489879 | closed | 0 | 11 | 2018-07-25T21:55:47Z | 2023-03-29T16:01:06Z | 2023-03-29T16:01:05Z | NONE | Looking for guidance on best practices, or forcing dask/xarray to work in the way I want it to work :) Task: I have hundreds of geoTIF files (500 for this test case), each containing a single 2d x-y array (~10000 x 10000). What I need to do is read these all in and apply a function to each x-y point whose argument is a 1-d array populated by the data from that same x-y point across all the files. This function is an optimization fit at each x-y point, so it's not something easily vectorizable, but each point's calculation is independent. Because each point may be treated independently, the idea is to be able to split the dataset into chunks in the x-y directions and allow Dask to cycle through and fork out the tasks of applying the function to each chunk in turn. Here's how I'm currently trying to implement this: ```python import xarray as xr import numpy as np Get list of files, set chunksizefilelist = ['image_001.tif', 'image_002.tif', ... 'image_500.tif'] chunksize=300 # Have explored a range of chunksizes from 250-1000 Open rasters individually in the absence of an open_mfdataset equivalent for rasterioAlternatively tried chunking or not in x and y dimensionsChunking at this stage blows up the task graph and eats up memory quicklybut see expected behavior below for reasoning behind trying to chunk at this stagerasterlist = [xr.open_rasterio(x, chunks={'x': chunksize, 'y': chunksize}) for x in filelist]rasterlist = [xr.open_rasterio(x, chunks={'x': None, 'y': None}) for x in filelist] Concatenate along band dimensionmerged = xr.concat(rasterlist, dim='band') Rechunkmerged = merged.chunk({'band': None, 'x': chunksize, 'y': chunksize}) Use dask map_blocks to apply a function to each chunk. This triggers the compute.output = merged.data.map_blocks(myfunction, dtype=np.float32, drop_axis=0) output = output.compute() ``` Problem descriptionWhat happens now is that all the raster datasets appear to be read into memory in full immediately and without respect to the final chunk alignment needed for the calculation. Then, there seems to be a lot of memory usage and communications overhead as the different chunks get rectified in the concatenation and merge-rechunk operations before myfunction is applied. This blows up the memory requirements, exceeding 300 GB RAM on a 48 worker machine (dataset on disk is ~100 GB). Below is a screenshot of the Dask dashboard right before workers start cutting out with memory limits/can't dump to disk errors.
Expected OutputWhat I'm looking for in this is an out-of-core computation where a single worker stays with a single chunk of the data all the way through to the end. Each "task" would then be a sequence of: - Open each of the raster input files and extract only the spatial chunk we were assigned for this task - Concatenate these for the computation - Perform this computation on this chunk - Return the output array for this chunk to be aggregated back together by Dask - Release data from memory; move on to the next spatial chunk I could manually sort this all out with a bunch of dask.delayed tasks, keeping track of the indices/locations of each chunk as they were returned to "reassemble" them later. However, is there an easier way to simplify this operation directly through xarray/dask through some ordering of commands I haven't found yet? Does the netcdf open_mfdataset function somehow handle this sort of idea for multiple netcdf files that could be adapted for rasterio? Output of
|
{
"url": "https://api.github.com/repos/pydata/xarray/issues/2314/reactions",
"total_count": 0,
"+1": 0,
"-1": 0,
"laugh": 0,
"hooray": 0,
"confused": 0,
"heart": 0,
"rocket": 0,
"eyes": 0
} |
completed | 13221727 | issue |